
Description
DeepHiC: A generative adversarial network for enhancing Hi-C data
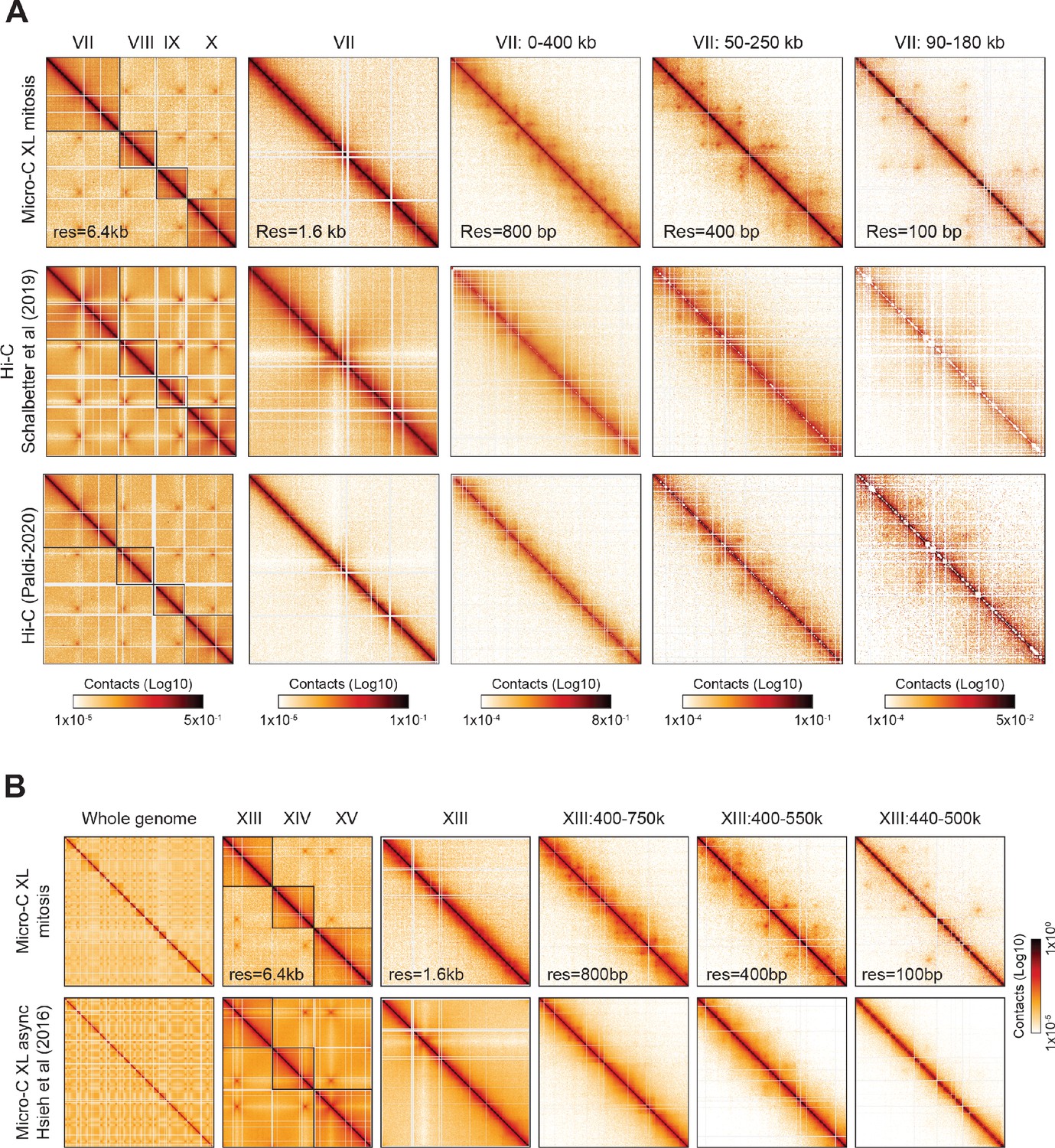
Cohesin residency determines chromatin loop patterns

Suhn K. Rhie's research works University of Southern California, California (USC) and other places

Comparison of the Hi-C, GAM and SPRITE methods using polymer
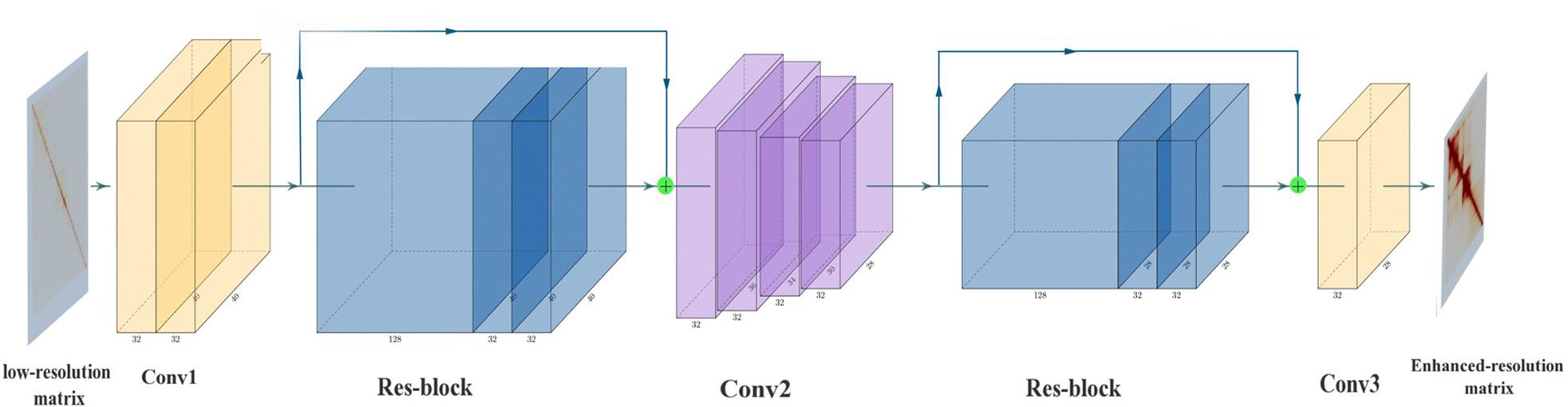
Frontiers SRHiC: A Deep Learning Model to Enhance the Resolution

Comparison with 3C-based assays a, Schematics of Hi-C, CHi-C, and

PDF) Characterizing chromatin interactions of regulatory elements and nucleosome positions, using Hi-C, Micro-C, and promoter capture Micro-C

HiCcompare: an R-package for joint normalization and comparison

Comparison of computational methods for 3D genome analysis at
Related products
You may also like

Sexy Panties for Women Cute Funny Underwear Naughty 3D Printed Animal Tail Underwears Briefs Gifts with Cute Ears, B-black, One Size : : Clothing, Shoes & Accessories

Winter Running Layers Guide for Cold Weather

Köp Lounge Underwear Intimates Set 30G På Nätet - Lounge Underwear Sverige Rea

Magazine Women's Racerback Bras Seamless Comfy Wireless Sports Bra
$ 8.00USD
Score 4.5(348)
In stock
Continue to book
You may also like

Sexy Panties for Women Cute Funny Underwear Naughty 3D Printed Animal Tail Underwears Briefs Gifts with Cute Ears, B-black, One Size : : Clothing, Shoes & Accessories

Winter Running Layers Guide for Cold Weather

Köp Lounge Underwear Intimates Set 30G På Nätet - Lounge Underwear Sverige Rea

Magazine Women's Racerback Bras Seamless Comfy Wireless Sports Bra
$ 8.00USD
Score 4.5(348)
In stock
Continue to book
©2018-2024, farmersprotest.de, Inc. or its affiliates



