GitHub - tgac-vumc/ACE: Absolute Copy Number Estimation using low-coverage whole genome sequencing data
Absolute Copy Number Estimation using low-coverage whole genome sequencing data - tgac-vumc/ACE

Assessing Copy Number Alterations in Targeted, Amplicon-Based Next-Generation Sequencing Data - ScienceDirect
awesome-omics/GENOMICS.md at main · flowhub-team/awesome-omics · GitHub

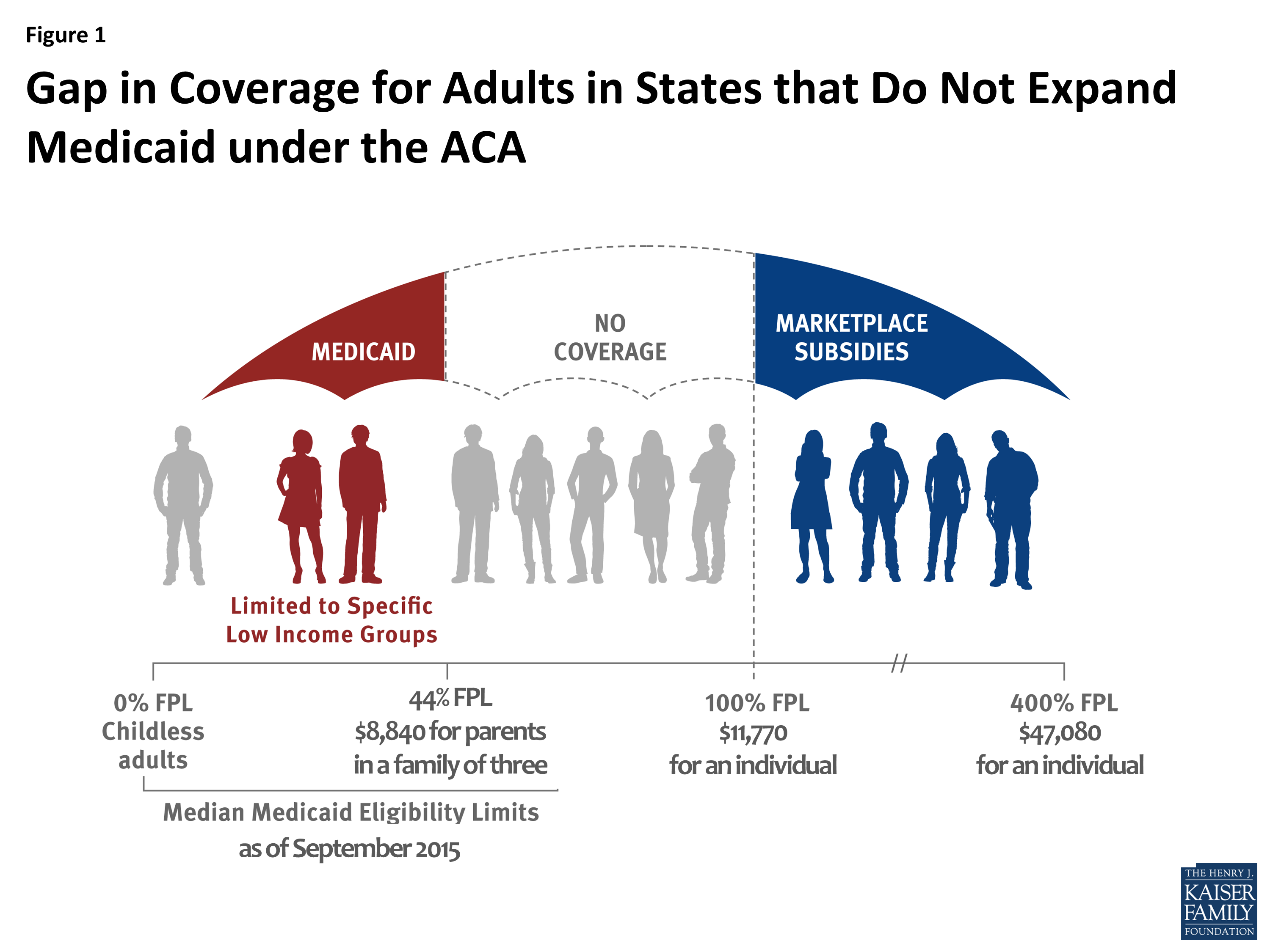
Absolute copy number fitting from shallow whole genome sequencing data
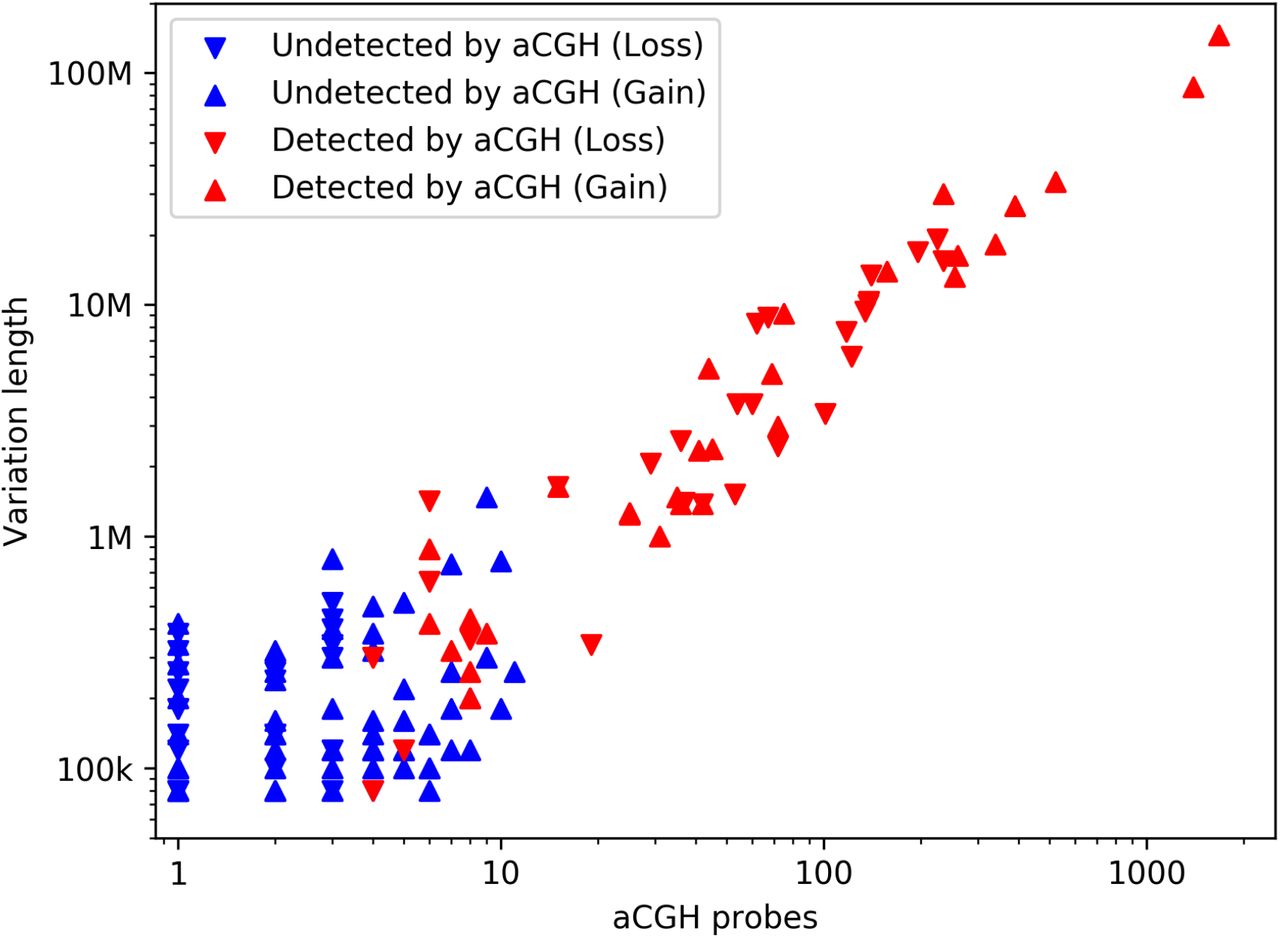
Copy number variant detection with low-coverage whole-genome sequencing is a viable alternative to the traditional array-CGH

Detection of Copy Number Variation using Shallow Whole Genome Sequencing Data to replace Array-Comparative Genomic Hybridization Analysis
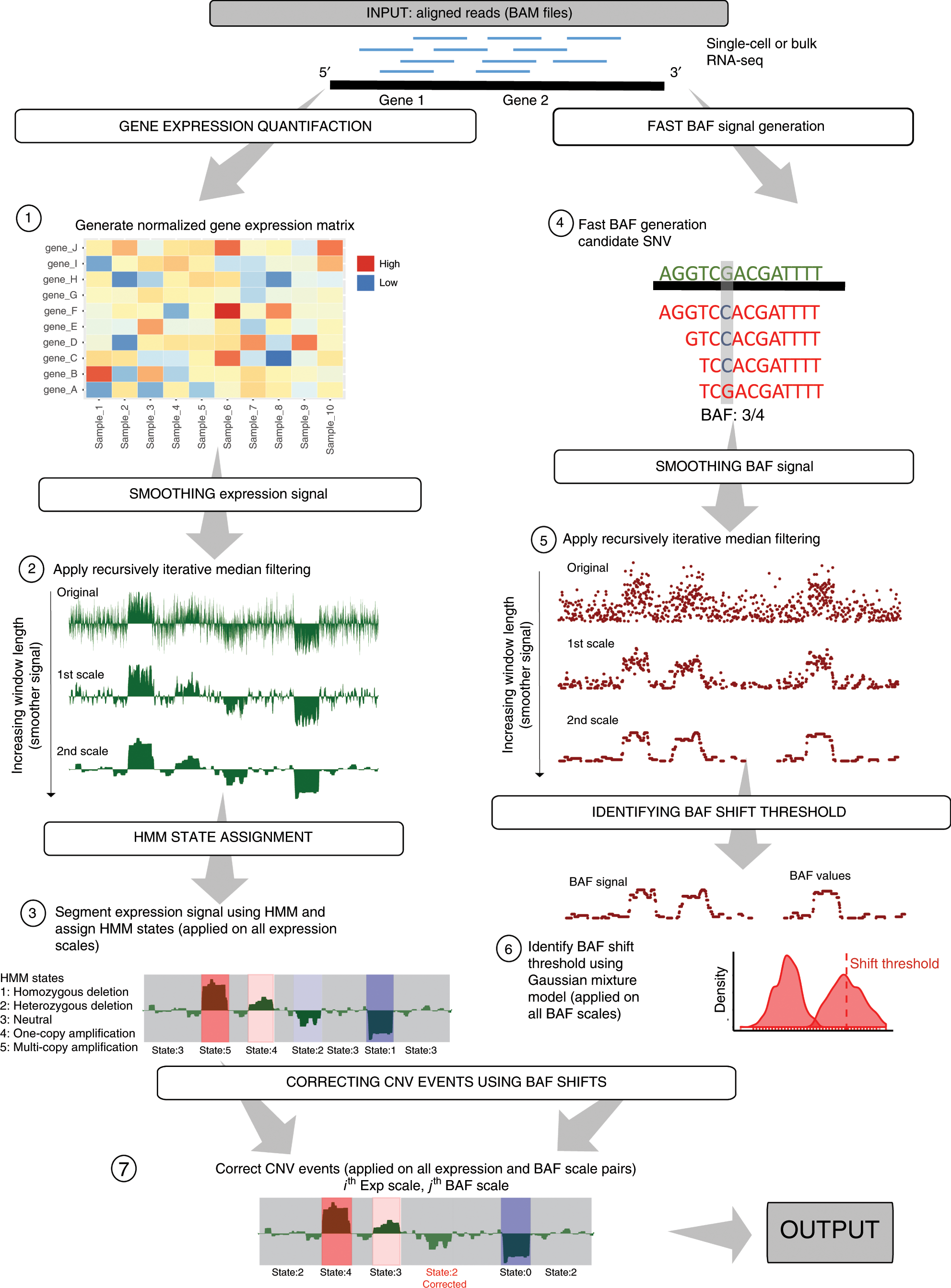
CaSpER identifies and visualizes CNV events by integrative analysis of single-cell or bulk RNA-sequencing data
GitHub - DKFZ-ODCF/ACEseqWorkflow: Allele-specific copy number estimation with whole genome sequencing
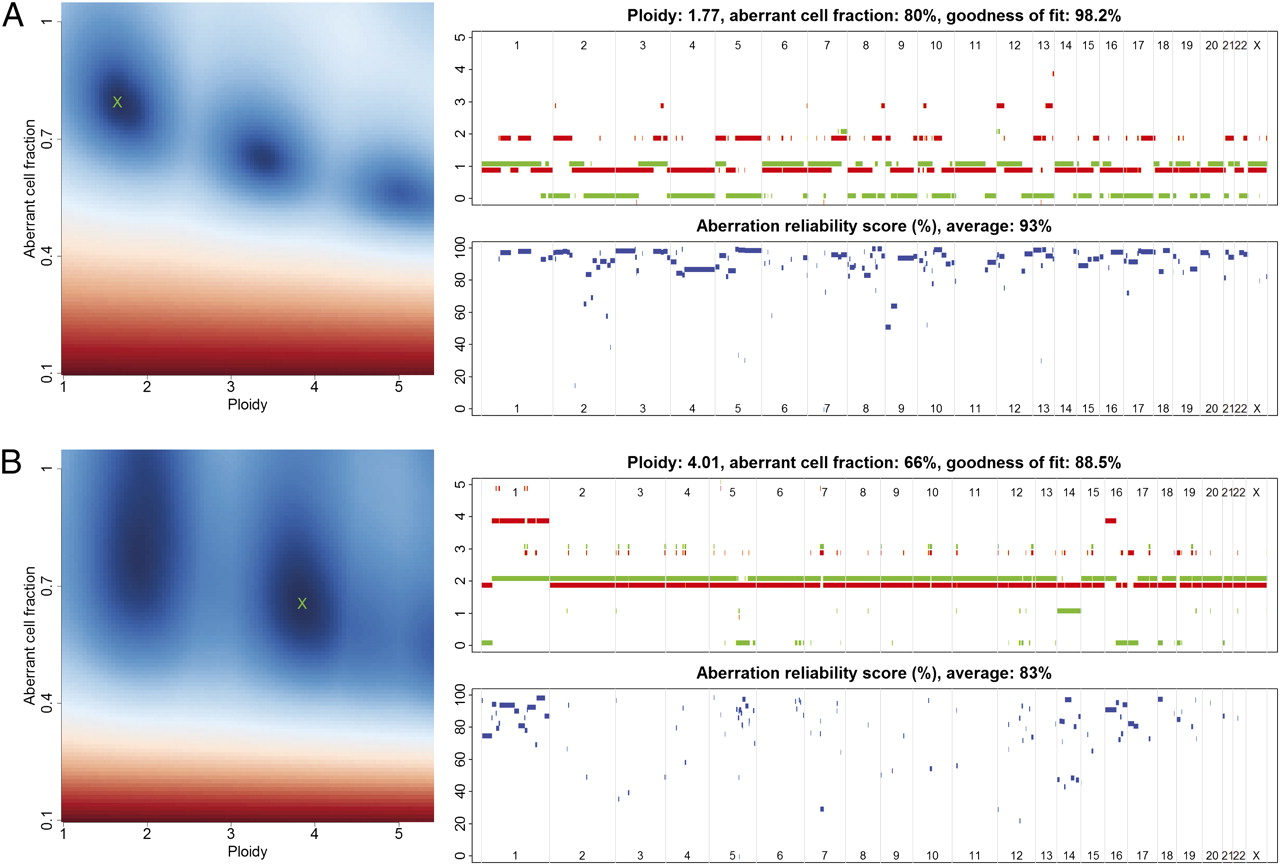
OUH - Cancer Genome Variation

PDF) HBOS-CNV: A New Approach to Detect Copy Number Variations From Next-Generation Sequencing Data

RNAseqCNV: analysis of large-scale copy number variations from RNA-seq data

CODEX2: full-spectrum copy number variation detection by high-throughput DNA sequencing, Genome Biology

Accurate quantification of copy-number aberrations and whole-genome duplications in multi-sample tumor sequencing data